In 1953, the discovery of the double helix structure of the DNA molecule was first announced. 60 years since that breakthrough discovery, scientists have continuously strived to develop gene sequencing technology. Up to now, people have upgraded gene sequencing technology to the 3rd generation.
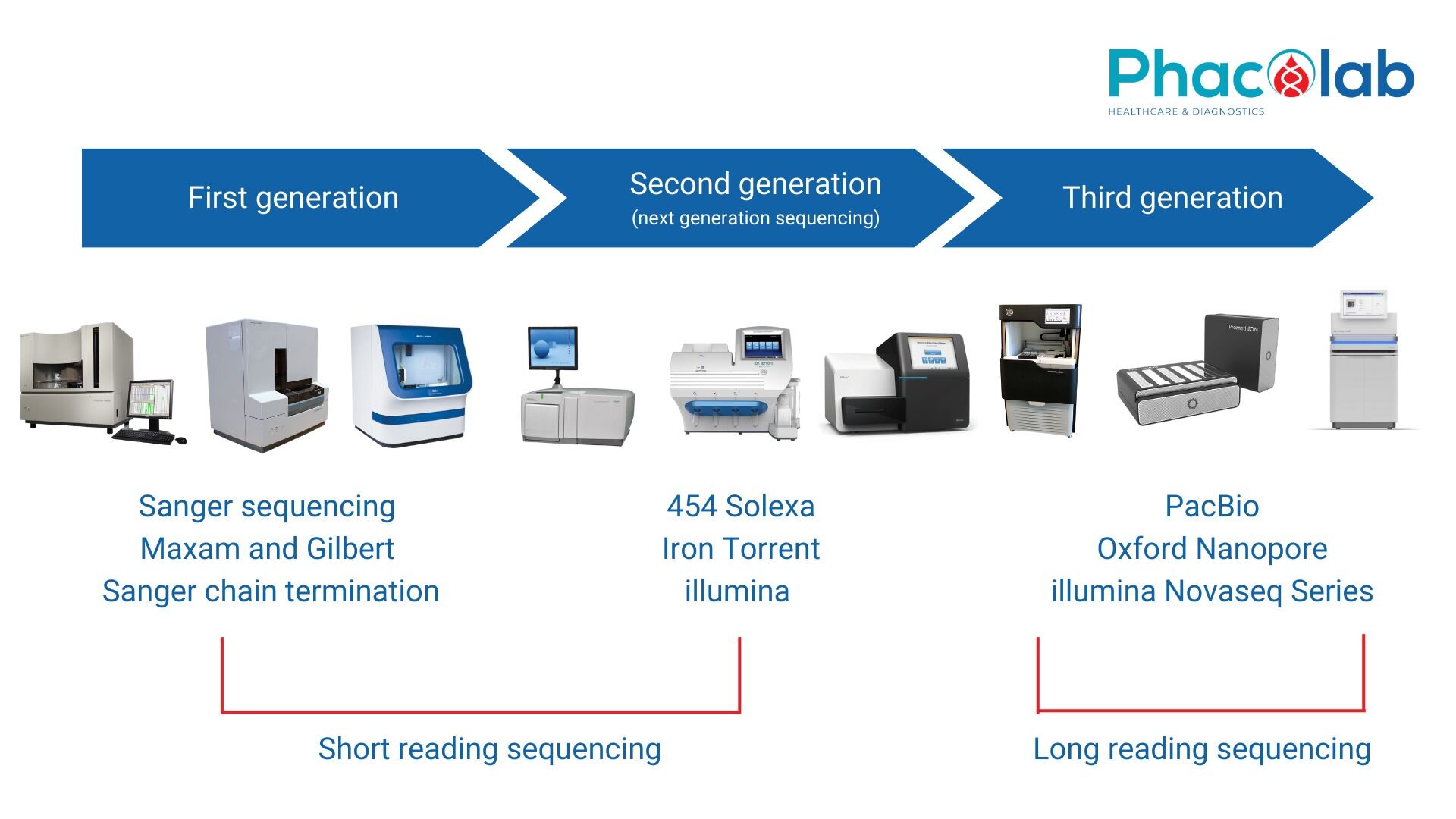
FIRST GENERATION SEQUING - THE BEGINNING OF THE ERA OF GENETICS
Sanger sequencing is the first generation DNA sequencing method, developed by Frederick Sanger and colleagues in 1977. In the 1980s, Sanger's original method was automated by scientists. studied at Caltech and commercialized by Applied Biosystems.
.
Four test tubes containing four different types of ddNTP labeled with radioactive isotopes were simultaneously subjected to a synthesis reaction. Then the products of 4 test tubes were electrophoresed and radioisotope imaging on the same gel to observe the DNA bands. Based on the position of the lines observed on the gel, determine the sequence of nucleotides in the direction from 5 to 3 from bottom to top.
.
Nowadays people use automatic sequencing machines to sequence electrophoretic bands. Dye temination sequencing uses fluorescently labeled ddNTPs to make DNA sequencing faster and more efficient. Each type of ddNTP is labeled with a different fluorescence emission wavelength, allowing sequencing to be performed in one reaction. Today, automated Sanger sequencing technology is still used, primarily in clinical laboratories where low product ratios, higher costs per sample, and long sequence reads of 500- 1,000 bp.
.
SECOND GENERATION SEQUING - SHORT READ BECOMES FAST AND EFFICIENT
Illumina, after acquiring Solexa company, has strongly developed new generation gene sequencing (NGS) technology. The key to NGS technology on Illumina's platform is "bridge amplification", which allows the formation of dense clusters of growth segments on a silicon chip. Sequencing is based on the principle of synthesis (SBS - Sequencing by synthesis), from the original single molecule is amplified into a large cluster of many copies, enough to detect the fluorescence signal when adding a single dNTP each time. Over time, the number of clusters that could be read simultaneously increased significantly, and Illumina's device became a successful commercialized mass parallel sequencing technology product. NGS is the sequencing technology mainly used today. Their maximum capacity allows sequencing at very low cost. However, NGS has a limit on read length; NGS platforms typically produce reads around 50-500 bp in length, suitable for resequencing projects, SNP calling, and targeted sequencing of very short amplicons.
.
THIRD GENERATION SEQUING - THE RISE OF LONG READ
Short reads are not suitable for all gene sequencing projects. PacBio's Single-molecule real-time sequencing (SMRT) technique is performed in a nano-sized hole with special properties: allowing observation of single-molecule fluorescence signals. when the fluorophore approaches the bottom of the nanohole (zero-mode waveguide).
.
Another long-read method based on molecular electrical signals was developed by Oxford Nanopore Technologies (ONT). The latest versions of single-molecule readout systems have been miniaturized significantly. Oxford Nanopore Technologies systems such as GridION, MinION or Flongle are handheld systems that can sequence RNA and DNA with read volumes of more than 2 Mb.
.
Illumina also launched the Novaseq series of instruments, reaching a new peak in sequencing power for commercial platforms. On each run, the NovaSeq 6000 (S4 flow cell) generates up to 3000 Gb of output (Illumina, 2017). Illumina's long-read devices are being perfected in the future, promising flexibility and increased scalability, helping to reduce the cost of sequencing the entire human genome.
.
.
Source:
https://www.genome.gov/dna-day https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4727787/
https://the-dna-universe.com/2020/11/02/a-journey-through-the-history-of-dna-sequencing/
https://www.pacb.com/blog/the-evolution-of-dna-sequencing-tools/





